
Primulina hunanensis K. M. Liu & X. Z. Cai is a perennial herb in the genus Primulina Hance of the family Gesneriaceae. It is very adaptable to low-light and barren cave environments, and is unique in its species evolution and environmental adaptation. P. hunanensis has beautiful flowers and a graceful shape that makes it valuable for horticultural cultivation. However, its habitat is unique and its distribution range is extremely narrow, occurring in only one county in Hunan Province. The wild population is estimated to be less than 800 plants, and it is classified as an endangered plant, representing a very small population of wild plants in Hunan.
Mitochondrial genome (mtDNA) research has also been applied to plant conservation and genetic improvement. In Gesneriaceae, mtDNA of only two species has been reported.
Researchers from the Wuhan Botanical Garden of the Chinese Academy of Sciences, in collaboration with researchers of Hunan Normal University, completed the sequencing and assembly of the complete mtDNA of P. hunanensis for the first time by using the latest sequencing technology with the advantage of long-reads (Nanopore).
The researchers analyzed the mtDNA features of P. hunanensis, including gene content, codon usage preference, repetitive sequences, RNA editing, and comparative phylogenetic relationships and genome covariance with the published mtDNA in Labiatae.
They found that the mtDNA of P. hunanensis was 575,242 bp in total length, with a GC content of 43.54%. Long-read data supported the assembly into a linear structure encoding a total of 60 genes, including 37 protein-coding genes, 20 transfer RNA genes, and three ribosome RNA genes.
In addition, protein-coding genes accounted for 9.32% of the total mtDNA length, and a total of 31 codons had relative synonymous codon usage values greater than 1.116 simple repeats, seven tandem repeats, and 362 dispersed repeat sequences were detected, and 455 potential RNA editing sites were identified.
Covariance results indicated a large number of genomic rearrangements between P. hunanensis and closely related species. The topology of the mitochondrial genome-based phylogeny showed that P. hunanensis was closely related to Boea hygrometrica.
This is the first complete mtDNA within Primulina and the third complete mtDNA within Gesneriaceae to be reported, which is an important addition to the limited plant mtDNA database.
The results were published in BMC Genomics titled "Assembly and comparative analysis of the initial complete mitochondrial genome of Primulina hunanensis (Gesneriaceae): a cave-dwelling endangered plant."
This study was supported by the National Wild Plant Germplasm Resource Center of China, the National Science & Technology Fundamental Resources Investigation Program of China, and the National Natural Science Foundation of China, etc.
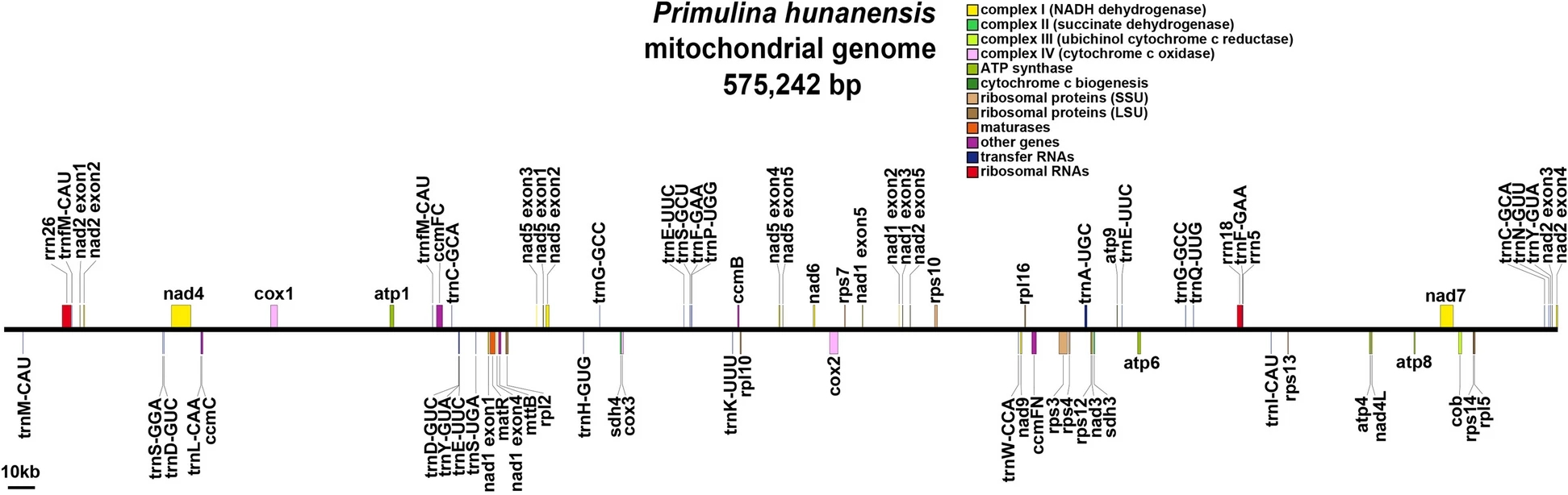
MtDNA map of P. hunanensis. (Image by WBG)
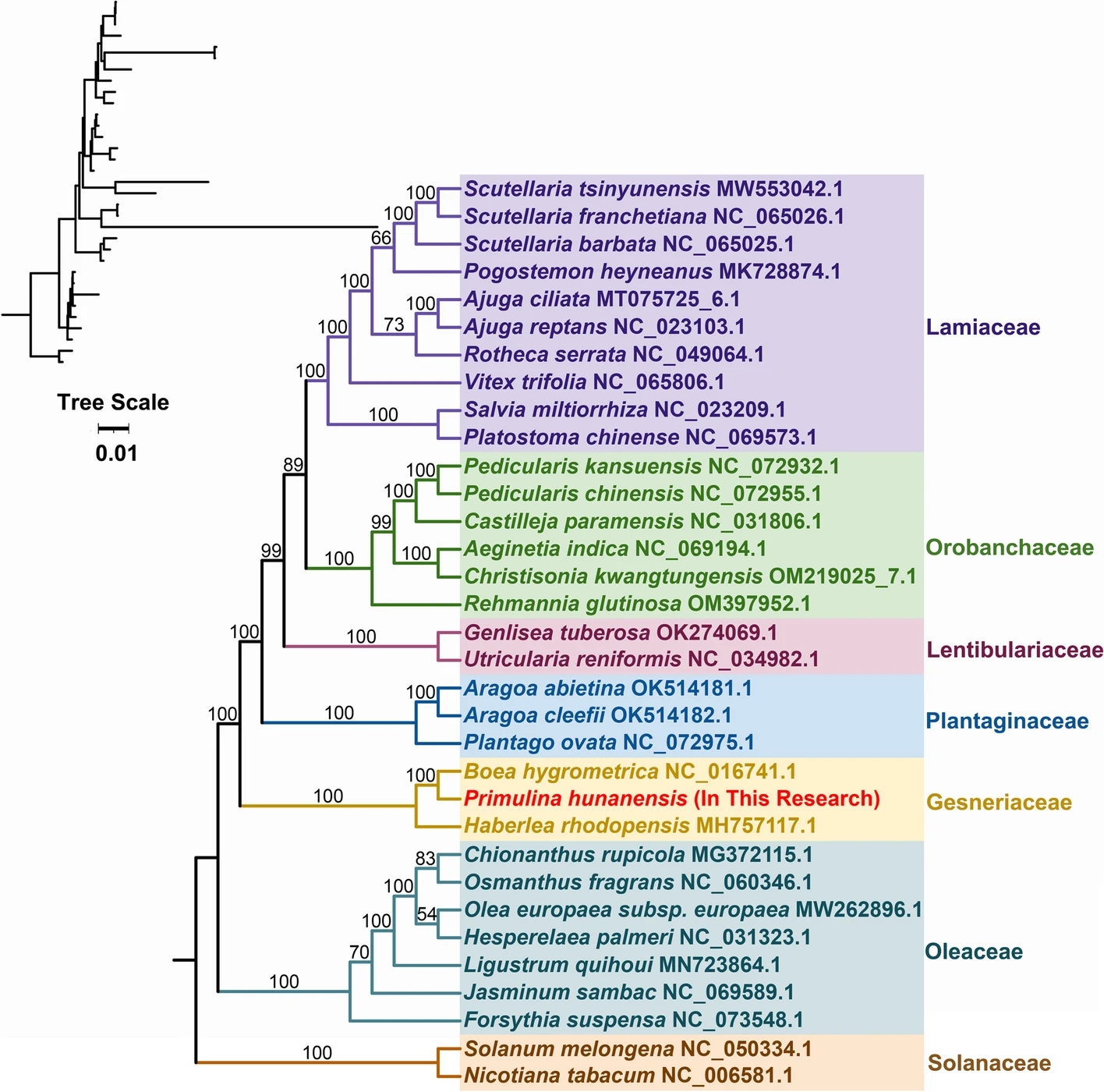
The phylogenetic associations of P. hunanensis alongside 32 other plant species. Phylogenetic tree constructed by maximum likelihood method based on 26 conserved mitochondrial genomic PCGs. (Image by WBG)

86-10-68597521 (day)
86-10-68597289 (night)

86-10-68511095 (day)
86-10-68512458 (night)

cas_en@cas.cn

52 Sanlihe Rd., Xicheng District,
Beijing, China (100864)

